DESCRIPTION
SPECIFICATION
RESOURCES

Service Description
We have developed innovative solutions for optimizing the process of accurately quantifying proteins in complex biological matrices and other complex mixtures.
Isobaric Label TMT Quantitation Proteomics
Isobaric Label services incorporate sample multiplexing to provide deep proteome coverage and highly precise quantitation of small or medium-sized sample sets. Samples are prepared and labeled separately, then all samples are pooled and analyzed by nano-flow LC-MS/MS1.
Project Workflow
BGI Genomics provides Isobaric Label Quantitative Proteomics service using TMT (Tandem Mass Tags for Quantitation) technology developed by Thermo Fisher Scientific.
- Protein extraction
- Protein digestion
- Isobaric labeling
- HPLC fractionation
- LC-MS/MS (DDA)
- Data analysis
Reference
[1] Wong, J. W. H., & Cagney, G. (2009). An Overview of Label-Free Quantitation Methods in Proteomics by Mass Spectrometry. Proteome Bioinformatics, 273–283.doi:10.1007/978-1-60761-444-9_18.
How to order
Mass Spectrometry Service Specification
Isobaric Label Quantitative Proteomics services are performed using nano-flow liquid chromatography and cutting-edge mass spectrometer (Q Exactive HF/HF-X).

Sample Preparation and Services
- The QC of enzymatic hydrolysis and labeling steps are strictly controlled by LC-MS DDA analysis
- 1h/fraction; 20 fractions for eukaryotes;10 fractions for prokaryotes

Quality Standard
- Summary includes all methods and data analysis
- Reports provided in Excel or PDF format, RAW files available upon request

Turn Around Time
- Typical 4-5 weeks from sample QC acceptance to data report delivery
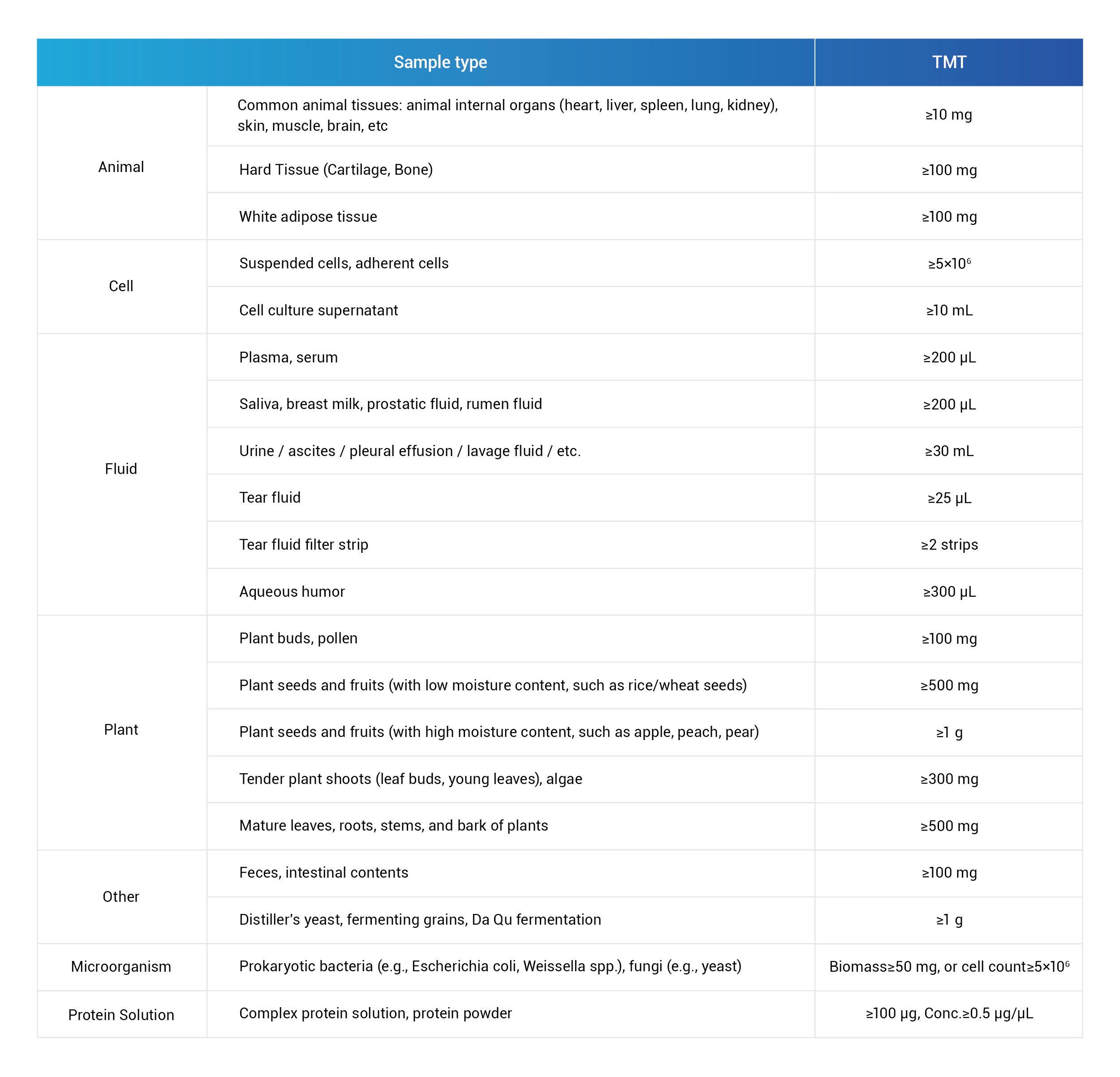
Sample Requirements
Data Analysis
DATA ANALYSIS
- Protein identification and quantification list
- Differential proteins data statistics and volcano plot
- Principal component analysis (PCA) and cluster analysis
- Protein GO/COG/KOG/Pathway annotation
- GO/COG/KOG enrichment analysis of differential proteins
- Protein-protein interaction analysis
- Protein subcellular localization analysis
- Repeatability analysis
Resources
We care for your samples from the start through to the result reporting. Highly experienced laboratory professionals follow strict quality procedures to ensure the integrity of your results.


